Introduction
Polyploidy refers to the presence of more than two sets of chromosomes within a single organism (Mason 2017). Polyploidy plays important roles in agriculture (Hilu 1993), species evolution (Otto and Whitton 2000; Joly et al. 2006; Wendel 2000), medicine (Coward and Harding 2014), among other fields. The research on polyploids is thus extensive. Several agricultural crops, such as wheat, soybean, potatoes, sugarcane, and cotton are polyploids (Hilu 1993), and polyploidy plays an important role on plant evolution (Otto and Whitton 2000). Polyploidization is also a property of some cancerous cells (Coward and Harding 2014). Therefore, studying polyploids is of great significance.
Polyploids are often divided into two categories based on their evolutionary histories — allopolyploids and autopolyploids (Cao, Osborn, and Doerge 2004). Autopolyploids arise from whole genome duplications within a single species. Allopolyploids arise by hybridization between species yielding polyploidization (Bourke et al. 2017). Chromosomes are also divided into two categories. Two chromosomes that arise from the same species are called homologues. Two chromosomes that arise from separate species are said to be homoeologues.
Allopolyploids and autopolyploids are often distinguishable by their meiotic behavior, which exhibits greater complexity than in diploids. Meiosis is an elementary process in biology (Voorrips and Maliepaard 2012), which allows parents to send their genetic materials to the next generation. There are many steps to meiosis, but we will focus on the pairing behavior during prophase I. During prophase I, the nucleoli and nuclear envelop disappear, hom(oe)ologous chromosomes pair and exchange genetic material. In allopolyploids, homologous chromosomes always pair with each other during prophase I. In autopolyploids, hom(oe)ologous chromosomes randomly pair together with no preference.
The definition of autopolyploidy and allopolyploidy based on the
evolutionary background depends on arbitrary thresholds of evolutionary
relatedness of homoeologues. Characterizing meiotic behavior requires no
such thresholds and so researchers have also suggested defining ploidy
by their meiotic behavior. However, unlike in diploids, where a parent
sends one copy to each offspring, in polyploids there is greater
diversity in the behavior of gametic transmission. For example, in
tetraploids (individuals with four copies of their genome) there are
Preferential pairing has been estimated in several ways. Ancestral origin frequencies are used to infer preferential pairing rates (Sybenga 1994). The method of haplotype reconstruction, provided in TetraOrigin, consists of multilocus parental linkage phasing and subsequent ancestral inference from SNP dosage data (Zheng et al. 2016). Preferential pairing can also be estimated by examining repulsion‐phase linkage patterns (Bourke et al. 2017).
Unfortunately, none of the above methods directly use genotype data, a common form of data in the modern sequencing age. For many types of data, the dosage of alleles are available at individual loci, but the phased configuration of these alleles are not known. These data often result from next-generation sequencing technologies that are then genotyped using genotyping software (Gerard et al. 2018; Gerard and Ferrão 2019; Clark, Lipka, and Sacks 2019; Blischak, Kubatko, and Wolfe 2018; Voorrips, Gort, and Vosman 2011). We use genotype data with F1 population (full-sib progeny of two parents) directly to analysis preferential pairing. Throughout, we assume that we are dealing with a tetraploid population and leave higher ploidy levels for future work. Therefore, In this thesis, we wish to distinguish between the types of ploidy and to estimate the strength of preferential pairing in polyploids from genotype data with F1 population directly.
In this thesis, we will use F1 population in genotype data, which consists of full-sib progeny of two parents. In an F1 population, each individual is the result of two meiotic divisions from each parent. We thus have replications of the meiotic behavior from each parent. We begin by providing a way to estimate preferential pairing at each locus from parents’ and progenies’ genotype. Due to the unidentifiability of which chromosomes contain which alleles, we provide a way based on clustering to identify this and estimate interpretable parameters. At last, through a simulation study, we demonstrate that our work can distinguish between the types of ploidy from genotype data directly.
The following is the outline of our paper. In Section 2.1, we provide a way to estimate preferential pairing at each locus using just genotype data. In Section 2.2, we provide a way to identify the parameterization and estimate interpretable parameters using model-based clustering. We demonstrate, through simulations, in Section 3.1 that we can accurately estimate the levels of preferential pairing at each locus through maximum likelihood estimation. We demonstrate in Section 3.2 that we can accurately distinguish between autopolyploidy and intermediate levels of polyploidy. However, we note that accurately detecting allopolyploidy is difficult. We conclude with a discussion in Chapter 4 .
Methods
Estimating Preferential Pairing using Maximum Likelihood Estimation
In this section, we will introduce a method to estimate preferential
pairing from genotype data. We will now describe the data under
consideration. We assume that we are working with an F1 population (a
biparental cross) which is common in breeding populations
(Sybenga 1994). An F1 population consists of parents and
offspring. We assume that we have genotype data from each parent and
each offspring. Genotype data consists of an integer
| Parameter | Means |
|---|---|
| The ploidy of the species. In our model, we assume all individuals are tetraploid (i.e. |
|
| The genotype of parent |
|
| Pairing configuration of a parent for a child |
|
| The number of copies of “aa” from parent |
|
| The number of copies of “Aa” from parent |
|
| The number of copies of “AA” from parent |
|
| The number of A’s sent by parent |
|
| The genotype of children |
|
| The probability of two chromosomes pairing together, |
|
| The probability of chromosome 1 and 2 pair together while chromosome 3 and 4 pair together. | |
| The probability of chromosome 1 and 3 pair together while chromosome 2 and 4 pair together. | |
| The probability of chromosome 1 and 4 pair together while chromosome 2 and 3 pair together. | |
| Preferential pairing parameter of parent |
To estimate preferential pairing, we need to understand the possible
pairing pattern in meiosis and define the preferential pairing
parameter. During prophase I, chromosomes will pair with each other. For
tetraploids, under bivalent pairing, there are three possible pairing
patterns: pair together, pair together, or pair together, which is
called a “pairing configuration”. To be more specific, we use
| |
|||||
|---|---|---|---|---|---|
| (2, 0, 0) | 1 | 0 | 0 | 0 | 0 |
| (1, 1, 0) | 0 | 1 | 0 | 0 | 0 |
| (1, 0, 1) | 0 | 0 | 0 | 0 | |
| (0, 2, 0) | 0 | 0 | 0 | 0 | |
| (0, 1, 1) | 0 | 0 | 0 | 1 | 0 |
| (0, 0, 2) | 0 | 0 | 0 | 0 | 1 |
After defining the parameter we interested, we need to find a method to
estimate the preferential pairing parameter. For tetraploids, during
prophase I, under bivalent pairing, there are three possible pairing
configurations: pairing together, pairing together, or pairing together.
During anaphase I and telophase I, pairing chromosomes move to the
opposite poles of the cell and separate into two individual cells.
Therefore, if the pattern is , one will be sent to the offspring.
Similarly, if the pattern is , one will be sent to the offspring.
However, if the pattern is , both and will have equal probability to be
sent to the offspring. We define
| (2, 0, 0) | 1 | 0 | 0 |
| (1, 1, 0) | 0.5 | 0.5 | 0 |
| (1, 0, 1) | 0 | 1 | 0 |
| (0, 2, 0) | 0.25 | 0.5 | 0.25 |
| (0, 1, 1) | 0 | 0.5 | 0.5 |
| (0, 0, 2) | 0 | 0 | 1 |
The above exposition results in a likelihood for the preferential
pairing parameter,
Combining information between loci to identify preferential pairing parameters via model-based clustering
In the previous section, we obtained preferential pairing parameter
estimates (the
Clustering by Stan
During prophase I, tetraploids, which contain four copies of chromosomes
in their genomes, will pair bivalently, and there are three possible
pairing behaviors. The four copies are denoted as chromosomes 1, 2, 3,
and 4. Under bivalent pairing, there are three possible formations: (i)
1 and 2 pair together while 3 and 4 pair together; (ii) 1 and 3 pair
together while 2 and 4 pair together; and (iii) 1 and 4 pair together
while 2 and 3 pair together. Let
Standard results from maximum likelihood theory guarantee asymptotic
normality of the maximum likelihood estimators
(Casella and Berger 2002). Therefore, we model the estimates
We will estimate the parameters in (3) using Bayesian approaches. This
requires setting priors over the parameters
where
To get the posterior probability
Note that, for
In order to obtain the posterior probability of each ploidy type, we
need to calculate the marginal distribution of the
Stan will provide posterior samples of the logs of
Theorem 2 For any given probability
Proof. For given probability
The proportion of autopolyploidy pairing behavior is
The proportion of intermediate pairing behavior is
The proportion of allopolyploidy pairing behavior is
Therefore, for given probability
If
If
we get $q^* = Pr(_b a^) = Pr(autopolyploidy|) +Pr(Intermediate|)Pr(_b<a^*|Intermediate) $.
ThenIf
Clustering by Dip Test
Since the problem of distinguishing between autopolyploidy and other
types of ploidy is equivalent to distinguishing between 1 and 2 clusters
of observations, we can explore more general methods to accomplish this
task. As our first comparison, we consider the dip test
(Hartigan, Hartigan, and others 1985). The dip test is used to test whether the given data
has more than one mode (Hartigan, Hartigan, and others 1985). The test statistic is called
the “dip” statistic, denoted as
Clustering by Mclust
As our second competitor, we will consider the Mclust R package
(Scrucca et al. 2016). Similar as dip test, Mclust can also distinguish
whether the given data has more than one mode. Mclust uses the normal
mixture model (17):
Results
Maximum Likelihood Simulation
To test our maximum likelihood estimation procedure in Section 2.1, we ran a simulation study. We varied the following parameters:
We tested each unique combination 1000 times, and got the MLE
The results are displayed in Figure 1. These are boxplots of
The accuracy is better as
The mean of the estimates is around the true value of
There is no obvious difference of estimates when
When the true value is near 0 or 1, the estimates are more accurate, when the true value is near 0.5, the estimates are relatively inaccurate.

Figure 1: Results of Maximu Likelihood Simulation
We therefore conclude that our method can accurately estimate
Simulation of model based on clustering
In Section 3.1 we demonstrated that we can consistently estimate
Data set
We used PedigreeSim to simulate datasets and evaluate our methods
(Voorrips and Maliepaard 2012). PedigreeSim is a software that generates simulated
genetic marker data of individuals in pedigreed populations. We used
PedigreeSim to generate an F1 population (full-sib progeny of two
parents) of individuals with varying levels of ploidy and preferential
pairing. In PedigreeSim we can vary some key parameters and build our
simulation. For example, we can set the ploidy of an organism (even
ploidy is supported), population type (selfings of one parent, full-sib
progeny of two parents, etc.), sample size, and so on. We also can vary
the properties of the chromosome, such as the length of the chromosome,
centromere positions, the amount of preferential pairing in polyploids,
from 0.0 (all pairwise combinations have equal probability, as in
autopolyploids) to 1.0 (fully preferential pairing, as in
allopolyploids), denoted as prefPairing, and the fraction of
quadrivalents, denoted as quadrivalents. Note that in PedigreeSim,
“prefPairing” is not the same as our definition of
We fixed the ploidy at 4 (tetraploids).
We set the the population type to F1: Full-sib progeny of two parents.
We varied sample size in
We set the length of chromosome to be 100
We set the location of the centromere to be at 50 cM, the midpoint of the chromosome
We set the preferential pairing parameter to vary in
We excluded quadrivalents and only allowed for bivalent pairing.
We fixed the number of SNPs to be 100.
We tested each unique combination of sample size and preferential
pairing parameter 100 times. For each iteration, we used PedigreeSim to
generate genotype data. Then we used the MLE method in Section
2.1 to estimate
Clustering results by Stan
After generating the genetic data and estimating the
| Autopolyploidy | 222 | 49 | 0 | 0 | 0 |
| Intermediate | 78 | 251 | 300 | 300 | 122 |
| Allopolyploidy | 0 | 0 | 0 | 0 | 178 |
| Accuracy | 74% | 83.7% | 100% | 100% | 59.3% |
Clustering results by Dip Test
Dip test can also help us to distinguish whether the data are from
autopolyploidy pairing behavior. Based on the model mentioned in Section
2.2.2, we cluster the
| Autopolyploidy | 286 | 229 | 120 | 10 |
| Non-autopolyploidy | 14 | 71 | 180 | 290 |
Clustering results by Mclust
Mclust is also a method which can help us to distinguish whether the
data are from autopolyploidy pairing behavior. Based on the model
mentioned in the section 2.2.3, we cluster the
| Autopolyploidy | 267 | 96 | 12 | 0 |
| Non-autopolyploidy | 33 | 204 | 288 | 300 |
Comparing three methods
To compare these three methods, we visualized the results in Figure
2, and display them in tabular form in Table
7. The plot shows the accuracy (with corresponding
95% confidence intervals based on exact binomial methods) for three
methods: Stan, Mclust, and dip test, under different levels of sample
size and the true value of
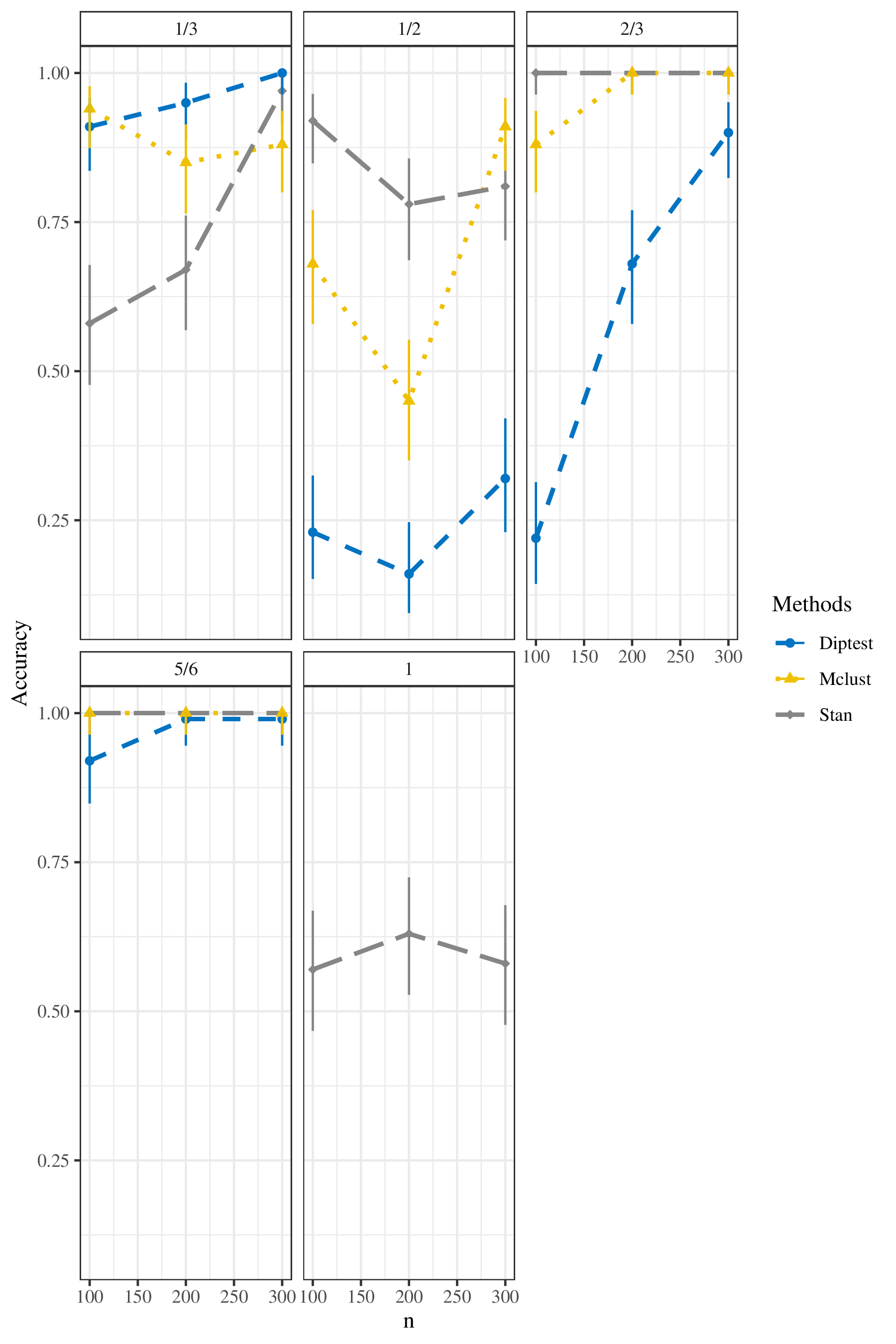 According to this plot, when the true value of
According to this plot, when the true value of
When the given data are from intermediate pairing behavior, the results
are slightly different. When the true value of
When the given data are from allopolyploidy pairing behavior, the true
value of
| Group | True |
Sample Size | Method | Accuracy | Lower | Upper |
|---|---|---|---|---|---|---|
| 1 | 100 | Dip Test | 0.91 | 0.84 | 0.96 | |
| 2 | 100 | Mclust | 0.94 | 0.87 | 0.98 | |
| 3 | 100 | Stan | 0.58 | 0.48 | 0.68 | |
| 4 | 200 | Dip Test | 0.95 | 0.89 | 0.98 | |
| 5 | 200 | Mclust | 0.85 | 0.76 | 0.91 | |
| 6 | 200 | Stan | 0.67 | 0.57 | 0.76 | |
| 7 | 300 | Dip Test | 1.00 | 0.96 | 1.00 | |
| 8 | 300 | Mclust | 0.88 | 0.80 | 0.94 | |
| 9 | 300 | Stan | 0.97 | 0.91 | 0.99 | |
| 10 | 100 | Dip Test | 0.23 | 0.15 | 0.32 | |
| 11 | 100 | Mclust | 0.68 | 0.58 | 0.77 | |
| 12 | 100 | Stan | 0.92 | 0.85 | 0.96 | |
| 13 | 200 | Dip Test | 0.16 | 0.09 | 0.25 | |
| 14 | 200 | Mclust | 0.45 | 0.35 | 0.55 | |
| 15 | 200 | Stan | 0.78 | 0.69 | 0.86 | |
| 16 | 300 | Dip Test | 0.32 | 0.23 | 0.42 | |
| 17 | 300 | Mclust | 0.91 | 0.84 | 0.96 | |
| 18 | 300 | Stan | 0.81 | 0.72 | 0.88 | |
| 19 | 100 | Dip Test | 0.22 | 0.14 | 0.31 | |
| 20 | 100 | Mclust | 0.88 | 0.80 | 0.94 | |
| 21 | 100 | Stan | 1.00 | 0.96 | 1.00 | |
| 22 | 200 | Dip Test | 0.68 | 0.58 | 0.77 | |
| 23 | 200 | Mclust | 1.00 | 0.96 | 1.00 | |
| 24 | 200 | Stan | 1.00 | 0.96 | 1.00 | |
| 25 | 300 | Dip Test | 0.90 | 0.82 | 0.95 | |
| 26 | 300 | Mclust | 1.00 | 0.96 | 1.00 | |
| 27 | 300 | Stan | 1.00 | 0.96 | 1.00 | |
| 28 | 100 | Dip Test | 0.92 | 0.85 | 0.96 | |
| 29 | 100 | Mclust | 1.00 | 0.96 | 1.00 | |
| 30 | 100 | Stan | 1.00 | 0.96 | 1.00 | |
| 31 | 200 | Dip Test | 0.99 | 0.95 | 1.00 | |
| 32 | 200 | Mclust | 1.00 | 0.96 | 1.00 | |
| 33 | 200 | Stan | 1.00 | 0.96 | 1.00 | |
| 34 | 300 | Dip Test | 0.99 | 0.95 | 1.00 | |
| 35 | 300 | Mclust | 1.00 | 0.96 | 1.00 | |
| 36 | 300 | Stan | 1.00 | 0.96 | 1.00 | |
| 37 | 1 | 100 | Stan | 0.57 | 0.47 | 0.67 |
| 38 | 1 | 200 | Stan | 0.63 | 0.53 | 0.72 |
| 39 | 1 | 300 | Stan | 0.58 | 0.48 | 0.68 |
Conclusion
The pairing behavior of tetraploid species was discussed in this thesis
and we described a way to directly estimate levels preferential pairing
at individual loci using genotype data derived from NGS data. From the
simulation study, we demonstrated that our model can estimate the
preferential pairing parameter
However, there are still a few weaknesses with our methods. By only exploring tetraploids, we excluded the consideration of higher ploidy levels. To avoid double reduction, the situation where one gamete receives two copies of part of the same parental hom(oe)olog, we only allow for bivalent pairing, and exclude quadrivalent pairing, which will lead to double reduction in some cases (Voorrips, Gort, and Vosman 2011; Bourke et al. 2017; Zheng et al. 2016).
Additionally, there are some statistical and computational limitations. For example, the number of iterations for simulation is relatively small, since we limited the number of simulation iterations for computational reasons. Additionally, as we mentioned above, MLE breaks down at the boundary, which leads to poor behavior when the given data are from allopolyploids. For future work, we will do more research to figure out the boundary issue (Marchand and Strawderman 2004; Zwiernik 2015).
Reference
Blischak, Paul D, Laura S Kubatko, and Andrea D Wolfe. 2018. “SNP Genotyping and Parameter Estimation in Polyploids Using Low-Coverage Sequencing Data.” Bioinformatics 34 (3): 407–15.
Bourke, Peter M., Paul Arens, Roeland E. Voorrips, G. Danny Esselink, Carole F. S. Koning-Boucoiran, Wendy P. C. van’t Westende, Tiago Santos Leonardo, et al. 2017. “Partial Preferential Chromosome Pairing Is Genotype Dependent in Tetraploid Rose.” The Plant Journal 90 (2): 330–43. https://doi.org/10.1111/tpj.13496.
Brent, Richard P. 1973. “Algorithms for Minimization Without Derivatives, Chap. 4.” Prentice-Hall Englewood Cliffs, NJ, USA.
Cao, Dachuang, Thomas C Osborn, and Rebecca W Doerge. 2004. “Correct Estimation of Preferential Chromosome Pairing in Autotetraploids.” Genome Research 14 (3): 459–62.
Casella, G., and R. L. Berger. 2002. Statistical Inference. Duxbury Advanced Series in Statistics and Decision Sciences. Thomson Learning. https://books.google.com/books?id=0x\_vAAAAMAAJ.
Clark, Lindsay V, Alexander E Lipka, and Erik J Sacks. 2019. “PolyRAD: Genotype Calling with Uncertainty from Sequencing Data in Polyploids and Diploids.” G3: Genes, Genomes, Genetics 9 (3): 663–73.
Coward, Jermaine, and Angus Harding. 2014. “Size Does Matter: Why Polyploid Tumor Cells Are Critical Drug Targets in the War on Cancer.” Frontiers in Oncology 4: 123.
Gerard, David, and Luís Felipe Ventorim Ferrão. 2019. “Priors for genotyping polyploids.” Bioinformatics 36 (6): 1795–1800. https://doi.org/10.1093/bioinformatics/btz852.
Gerard, David, Luís Felipe Ventorim Ferrão, Antonio Augusto Franco Garcia, and Matthew Stephens. 2018. “Genotyping Polyploids from Messy Sequencing Data.” Genetics 210 (3): 789–807. https://doi.org/10.1534/genetics.118.301468.
Hartigan, John A, Pamela M Hartigan, and others. 1985. “The Dip Test of Unimodality.” The Annals of Statistics 13 (1): 70–84.
Hilu, K. W. 1993. “Polyploidy and the Evolution of Domesticated Plants.” American Journal of Botany 80 (12): 1494–9. http://www.jstor.org/stable/2445679.
Joly, Simon, Julian Starr, Walter Lewis, and Anne Bruneau. 2006. “Polyploid and Hybrid Evolution in Roses East of the Rocky Mountains.” American Journal of Botany 93 (March): 412–25. https://doi.org/10.3732/ajb.93.3.412.
Maechler, Martin. 2016. Diptest: Hartigan’s Dip Test Statistic for Unimodality - Corrected. https://CRAN.R-project.org/package=diptest.
Marchand, Eric, and William E. Strawderman. 2004. “Estimation in Restricted Parameter Spaces: A Review.” Lecture Notes-Monograph Series 45: 21–44. http://www.jstor.org/stable/4356296.
Mason, Annaliese S. 2017. Polyploidy and Hybridization for Crop Improvement. CRC Press.
Otto, Sarah P, and Jeannette Whitton. 2000. “Polyploid Incidence and Evolution.” Annual Review of Genetics 34 (1): 401–37.
Scrucca, Luca, Michael Fop, T Brendan Murphy, and Adrian E Raftery. 2016. “Mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models.” The R Journal 8 (1): 289.
Stan Development Team. 2018. “Stan Modeling Language Users Guide and Reference Manual, Version 2.18.0.” http://mc-stan.org/.
———. 2020. “RStan: The R Interface to Stan.” http://mc-stan.org/.
Sybenga, J. 1994. “Preferential Pairing Estimates from Multivalent Frequencies in Tetraploids.” Genome 37 (6): 1045–55. https://doi.org/10.1139/g94-149.
Voorrips, Roeland E, Gerrit Gort, and Ben Vosman. 2011. “Genotype Calling in Tetraploid Species from Bi-Allelic Marker Data Using Mixture Models.” BMC Bioinformatics 12 (1): 172.
Voorrips, Roeland, and Chris Maliepaard. 2012. “The Simulation of Meiosis in Diploid and Tetraploid Organisms Using Various Genetic Models.” BMC Bioinformatics 13 (September): 248. https://doi.org/10.1186/1471-2105-13-248.
Wendel, Jonathan F. 2000. “Genome Evolution in Polyploids.” In Plant Molecular Evolution, 225–49. Springer.
Wu, Rongling, Maria Gallo-Meagher, Ramon C Littell, and Zhao-Bang Zeng. 2001. “A General Polyploid Model for Analyzing Gene Segregation in Outcrossing Tetraploid Species.” Genetics 159 (2): 869–82.
Zheng, Chaozhi, Roeland E Voorrips, Johannes Jansen, Christine A Hackett, Julie Ho, and Marco CAM Bink. 2016. “Probabilistic Multilocus Haplotype Reconstruction in Outcrossing Tetraploids.” Genetics 203 (1): 119–31.
Zwiernik, P. 2015. Semialgebraic Statistics and Latent Tree Models. Chapman & Hall/Crc Monographs on Statistics & Applied Probability. CRC Press. https://books.google.com/books?id=YdWYCgAAQBAJ.